The central goal of our research program is to understand how genetic information is transmitted accurately during cell division, a fundamental biological process that is critical for all life. We focus on the centromere, an essential chromosomal locus that directs kinetochore assembly and accurate chromosome segregation in mitosis and meiosis. The presence of one – and only one – centromere on each chromosome is required to prevent aneuploidy, chromosome breakage, and genome rearrangements, all of which are hallmarks of tumors, developmental defects, and infertility in humans. How a particular chromosomal region is selected as a centromere is poorly understood.
Despite their conserved function, centromeres are among the most rapidly evolving regions of genomes and their re-positioning through the formation of evolutionary new centromeres (ENCs) is an important driver of speciation. The centromeric DNA (cenDNA) of complex eukaryotes typically consists of large blocks of satellite DNA (satDNA) and interspersed transposable elements (TEs). The functions of satDNA and TEs are not well understood; however, recent studies suggest their transcription may be important for centromere function.
Despite their conserved function, centromeres are among the most rapidly evolving regions of genomes and their re-positioning through the formation of evolutionary new centromeres (ENCs) is an important driver of speciation. The centromeric DNA (cenDNA) of complex eukaryotes typically consists of large blocks of satellite DNA (satDNA) and interspersed transposable elements (TEs). The functions of satDNA and TEs are not well understood; however, recent studies suggest their transcription may be important for centromere function.
|
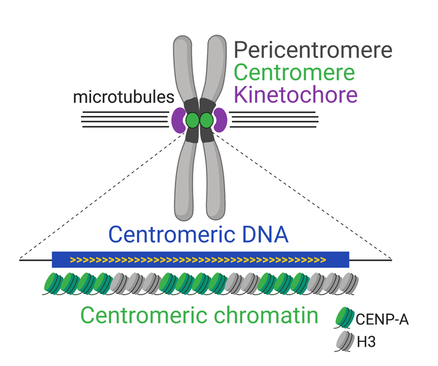
Figure 1. Function and organization of complex centromeres.
|
While cenDNA is not conserved at the sequence level, the presence of chromatin containing the centromere-specific histone CENP-A (Centromere Protein A; Figure 1) is nearly universal. CENP-A provides the structural cue for kinetochore assembly during cell division and, as such, is absolutely essential for viability. Even though CENP-A chromatin is faithfully maintained at the centromere through epigenetic inheritance, its de novo deposition at non-centromeric locations has been implicated in the formation of ENCs in mammals and neocentromeres in humans. Neocentromeres are associated with developmental defects and cancer. Thus, understanding how CENP-A is deposited at specific sites in the genome has far-reaching implications for evolution and human disease.
|
A long-standing model in the field stipulates that centromeres are specified epigenetically by CENP-A and do not require cenDNA. However, several observations suggest that cenDNA sequences may in fact be key centromere effectors, compelling a closer investigation of the contribution of CENP-A chromatin and cenDNA to centromere identity.
Our lab investigate centromere structure and function using Drosophila melanogaster (Dmel), an exceptional model for centromere biology. The centromeres of Dmel are smaller (100-170kb versus 630kb-1.8Mb) and fewer (four chromosomes versus 23) than those of humans and yet are similarly complex. Furthermore, Dmel offers unparalleled genetic tools, a wealth of genomic and phylogenetic resources, and the distinct advantage of enabling the analysis of centromere function in the context of the whole organism. We recently developed inducible de novo centromeres and complete centromere assemblies for this species, providing us with a powerful system to gain insights into how centromeres are specified.
Our lab investigate centromere structure and function using Drosophila melanogaster (Dmel), an exceptional model for centromere biology. The centromeres of Dmel are smaller (100-170kb versus 630kb-1.8Mb) and fewer (four chromosomes versus 23) than those of humans and yet are similarly complex. Furthermore, Dmel offers unparalleled genetic tools, a wealth of genomic and phylogenetic resources, and the distinct advantage of enabling the analysis of centromere function in the context of the whole organism. We recently developed inducible de novo centromeres and complete centromere assemblies for this species, providing us with a powerful system to gain insights into how centromeres are specified.
What is the role of DNA repeats in centromere identity?
Growing evidence points to a role for cenDNA in centromere specification. Our discovery of Dmel cenDNA in collaboration with the laboratory of Amanda Larracuente (Chang, Chavan, Palladino et al., PLoS Biol 2019) provides unprecedented access into metazoan centromere organization, laying the foundation for the manipulation of cenDNA in this powerhouse model organism.
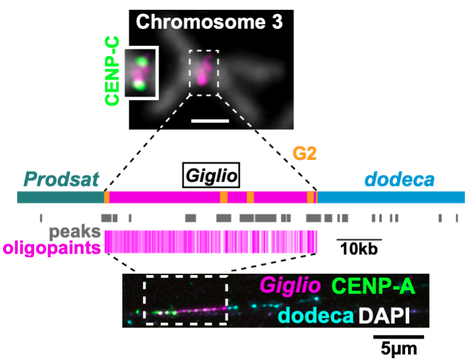
The Dmel centromeres consists of ‘islands’ of complex DNA enriched in retroelements (REs), a class of TEs, surrounded by large blocks of satDNA (Figure 2). Approximately 70% of the region occupied by CENP-A resides on the islands, the rest on satDNA. Each centromere island contains a distinct arrangement of REs and can be identified by OligoPaint Fluorescence In Situ Hybridization (FISH). Even though most are enriched at the centromere, all centromeric REs are also found elsewhere in the genome, and thus cannot be sufficient for centromere identity. However, REs may contribute to creating an optimal genomic landscape for centromere specification and propagation, perhaps through their transcription. Transcription of cenDNA has been implicated in centromere function in both a sequence-independent manner and through the formation of non-coding cenRNAs. Centromeric REs could provide a transcriptionally active chromatin environment to facilitate CENP-A deposition at each cell cycle or generate cenRNAs with structural roles in centromere function.
Growing evidence points to a role for cenDNA in centromere specification. Our discovery of Dmel cenDNA in collaboration with the laboratory of Amanda Larracuente (Chang, Chavan, Palladino et al., PLoS Biol 2019) provides unprecedented access into metazoan centromere organization, laying the foundation for the manipulation of cenDNA in this powerhouse model organism.
The Dmel centromeres consists of ‘islands’ of complex DNA enriched in retroelements (REs), a class of TEs, surrounded by large blocks of satDNA (Figure 2). Approximately 70% of the region occupied by CENP-A resides on the islands, the rest on satDNA. Each centromere island contains a distinct arrangement of REs and can be identified by OligoPaint Fluorescence In Situ Hybridization (FISH). Even though most are enriched at the centromere, all centromeric REs are also found elsewhere in the genome, and thus cannot be sufficient for centromere identity. However, REs may contribute to creating an optimal genomic landscape for centromere specification and propagation, perhaps through their transcription. Transcription of cenDNA has been implicated in centromere function in both a sequence-independent manner and through the formation of non-coding cenRNAs. Centromeric REs could provide a transcriptionally active chromatin environment to facilitate CENP-A deposition at each cell cycle or generate cenRNAs with structural roles in centromere function.
|
Figure 2. Centromere 3 organization. Top: mitotic chr3 from larval brain showing DNA FISH with OligoPaints for the cen3 island (Giglio). Inset, IF with the centromere marker CENP-C. Middle: diagram of cen3 contig showing Giglio with interspersed G2 REs and flanking satellites, the positions of uniquely mapping CENP-A ChIP-seq peaks and OligoPaints. Bottom: IF-FISH on extended chromatin fibers from larval brain showing the that CENP-A occupies primarily the island (1µm ≈10kb).
|
G2/Jockey-3 (G2) is the only RE shared amongst all centromeres and the most highly enriched repeat in CENP-A Chromatin Immunoprecipitations (ChIPs). Excitingly, G2 is highly enriched also at the centromeres of D. simulans, which harbors highly divergent centromeric satDNAs, suggesting a conserved role at the centromere. Identifying the functional significance of G2 and other REs in centromere identity is a major goal of our research program. Using a combination of genome editing, cell biological, and genomics approaches our lab is probing the role of the centromere islands, centromeric REs, and their transcription in centromere function, specification, and inheritance. We are also directly testing hypotheses to explain the conserved association between G2 and centromeres.
|
|
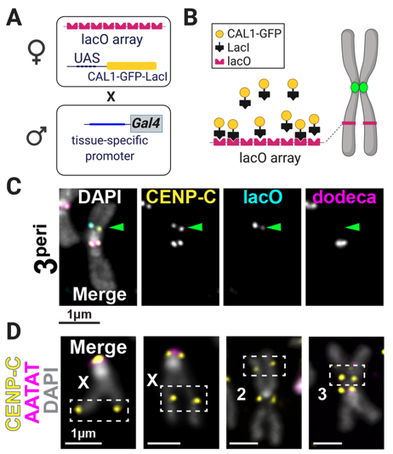
Figure 3. De novo centromere systems in flies. A Crosses for CENP-A deposition at lacO. B LacI/lacO schematic. C IF-FISH on larval brain mitotic chromosomes of progeny from A for pericentric lacO on Chr3. CENP-C (yellow) indicates active centromere; lacO (cyan); Dodeca satDNA (magenta). DAPI, gray. D IF-FISH on larval brain mitotic chromosomes in which CAL1 is overexpressed. Crosses are as in A, except that mothers do not contain a lacO array.
|
Is CENP-A chromatin sufficient for assembly and propagation of de novo centromeres in vivo?
If the epigenetic model of centromere identity is correct, CENP-A chromatin should specify centromere activity independently of whether it is assembled on cenDNA. However, the ability of centromeres specified by CENP-A alone to sustain centromere function long-term has not been tested in any multicellular organism. We devised the first in vivo system to investigate de novo centromere assembly and transmission through development in somatic cells (Palladino et al., Dev Cell, 2020). We generate de novo centromeres in two ways: 1) by targeting CENP-A chromatin to integrated lacO arrays at distinct genomic locations via LacI tethering of the CENP-A assembly factor CAL1, and 2) by inducing the formation of spontaneous de novo centromeres via CAL1 overexpression (Figure 3). Targeted de novo centromeres formed efficiently at six lacO positions representing diverse euchromatic and heterochromatic environments, indicating that chromatin is generally malleable to artificial conversion to a centromere. Spontaneous de novo centromeres formed at many different sites across the genome without any obvious sub-chromosomal preference. When we induced de novo centromeres early in embryogenesis to track their maintenance through development, we found that only those that caused less frequent chromosome breaks (i.e. pericentromeric) were efficiently maintained. Current projects in the lab are rigorously probing CENP-A sufficiency in vivo and investigating if spontaneous de novo CENP-A deposition occurs at preferential locations. |
Our research is currently funded by the NIH-NIGMS. We also acknowledge previous support from the NSF.